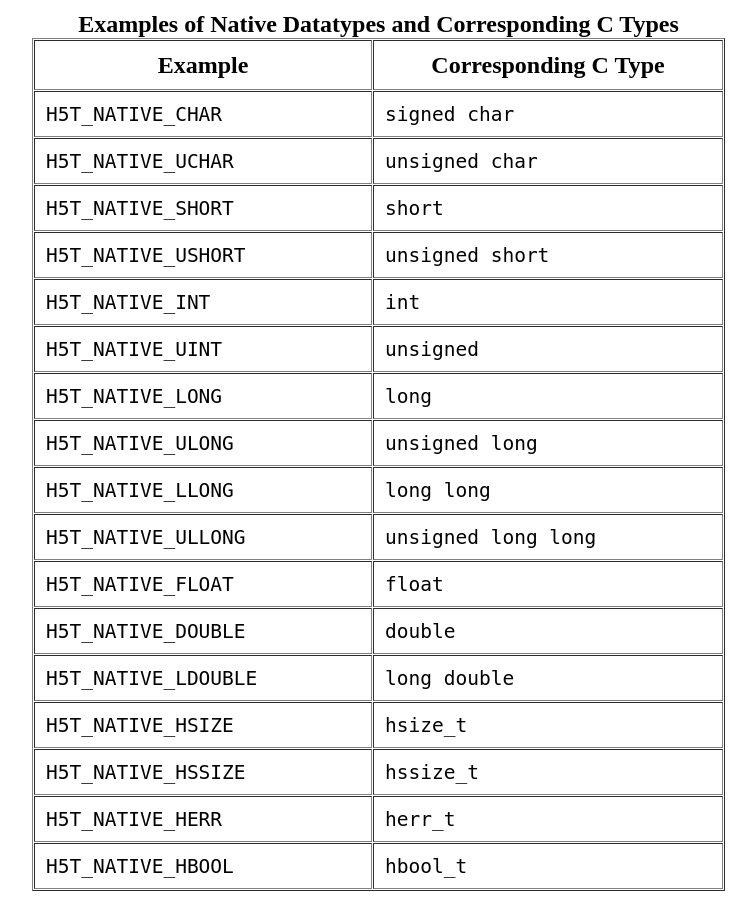
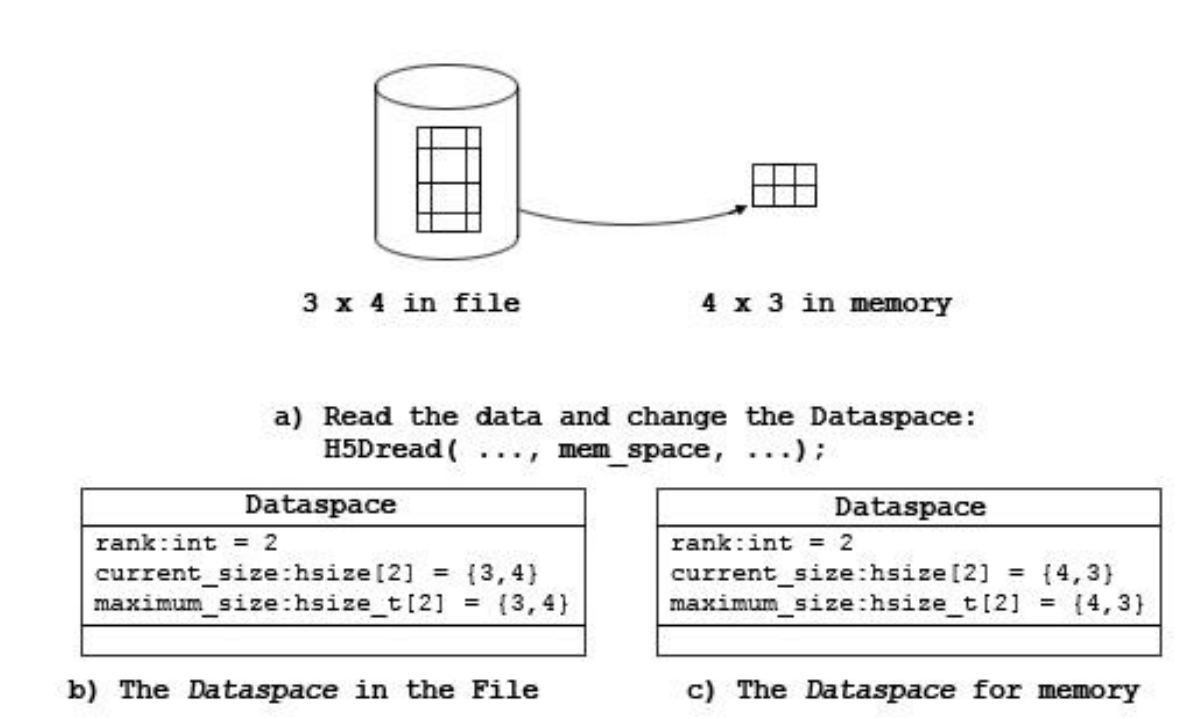
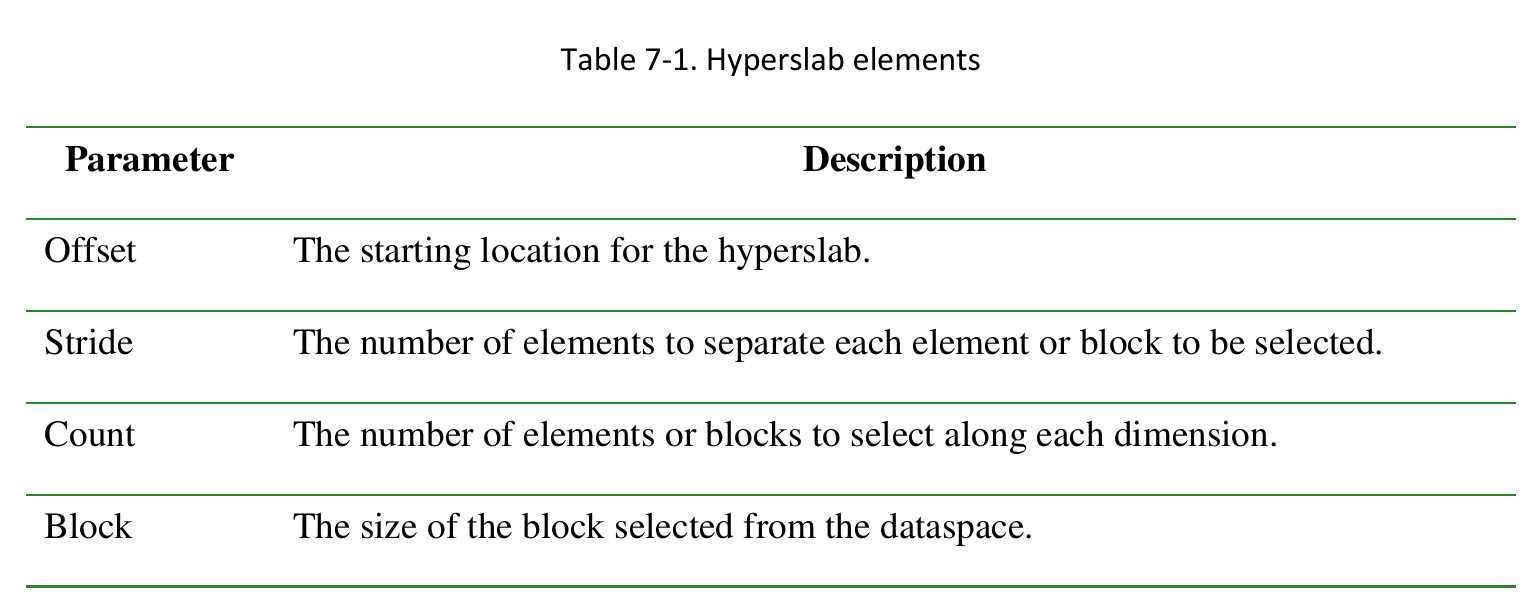
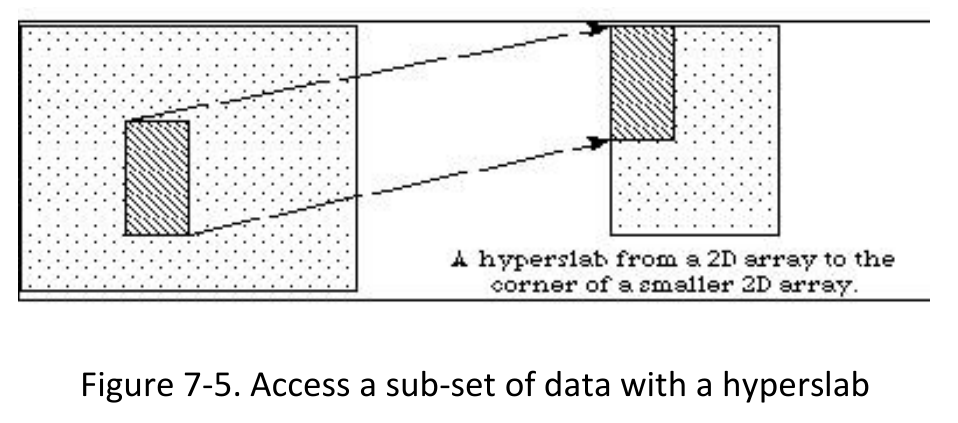
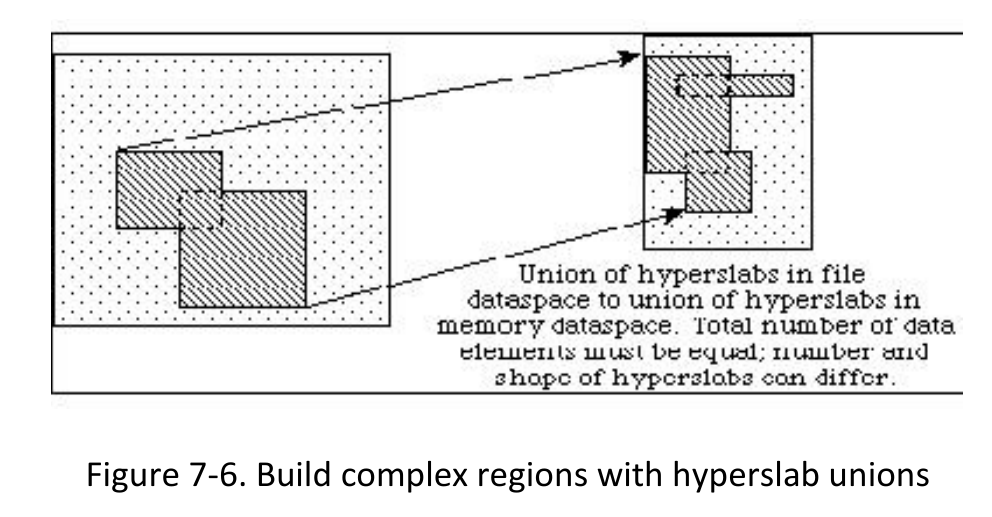
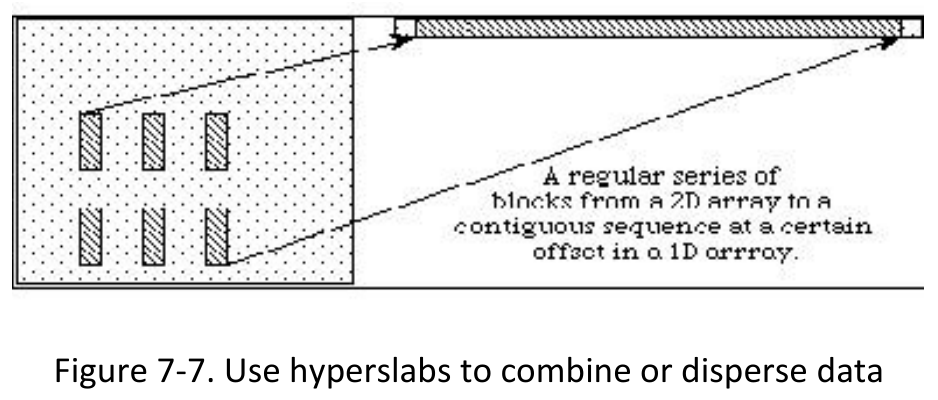
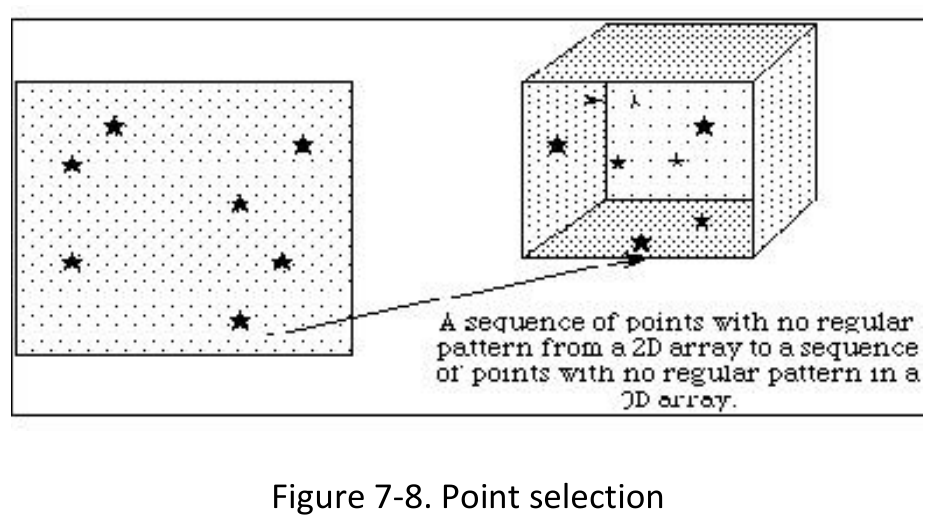
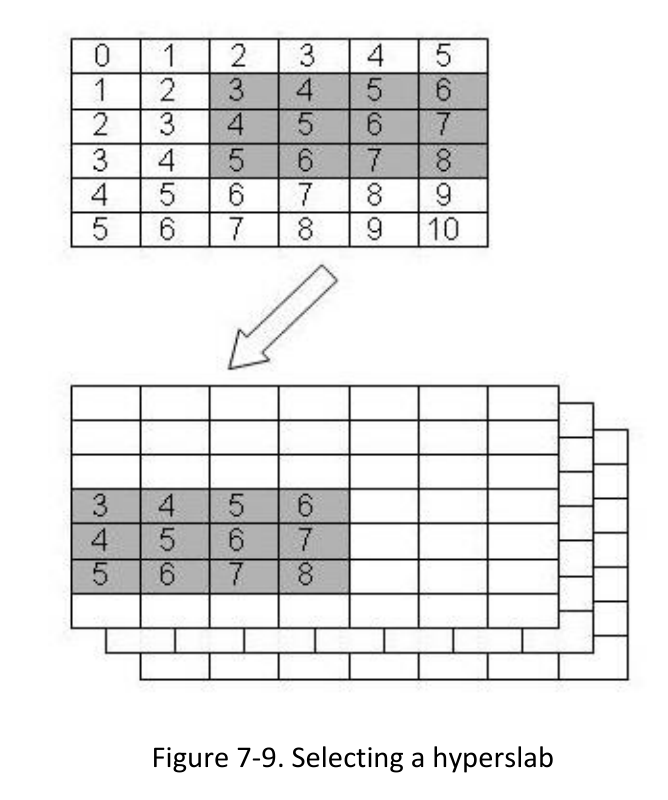
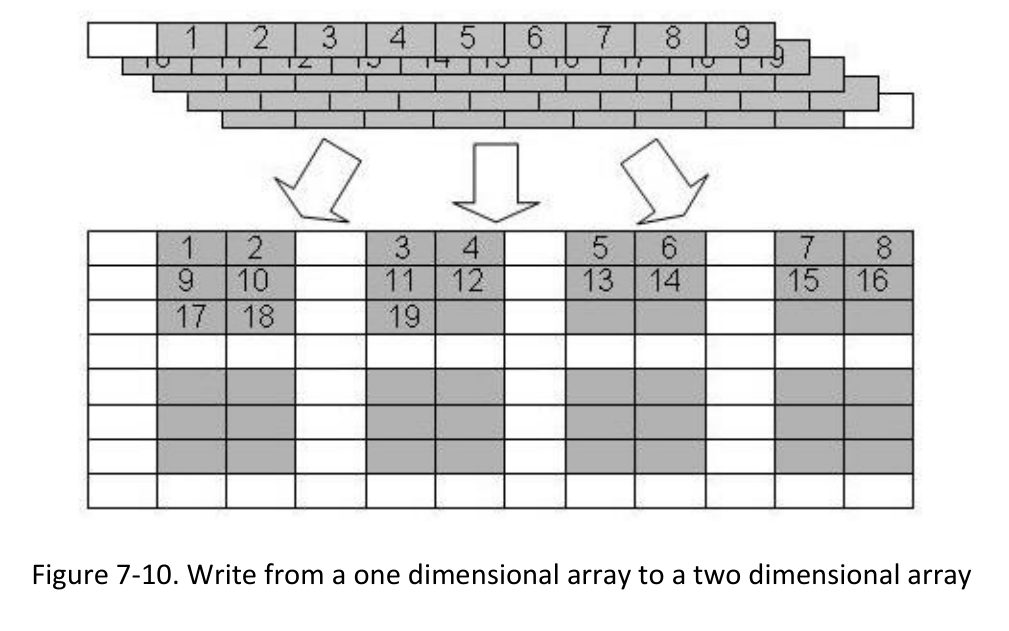
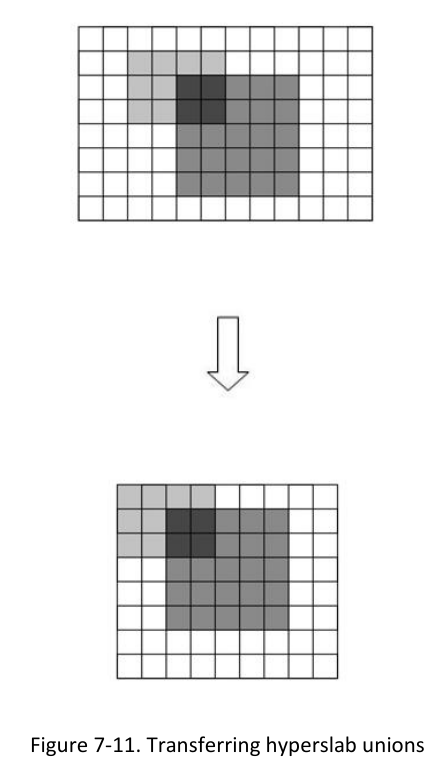
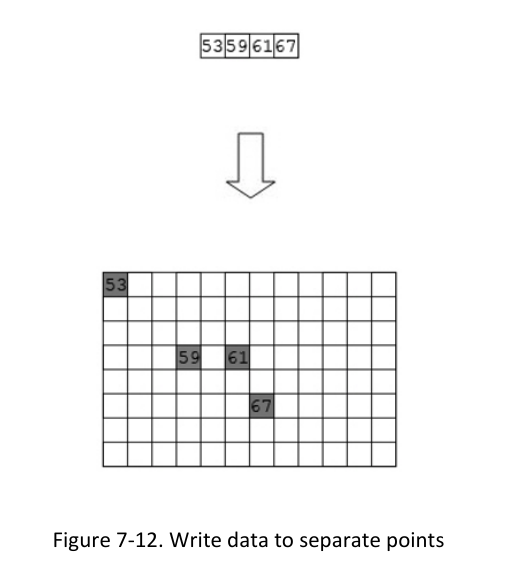
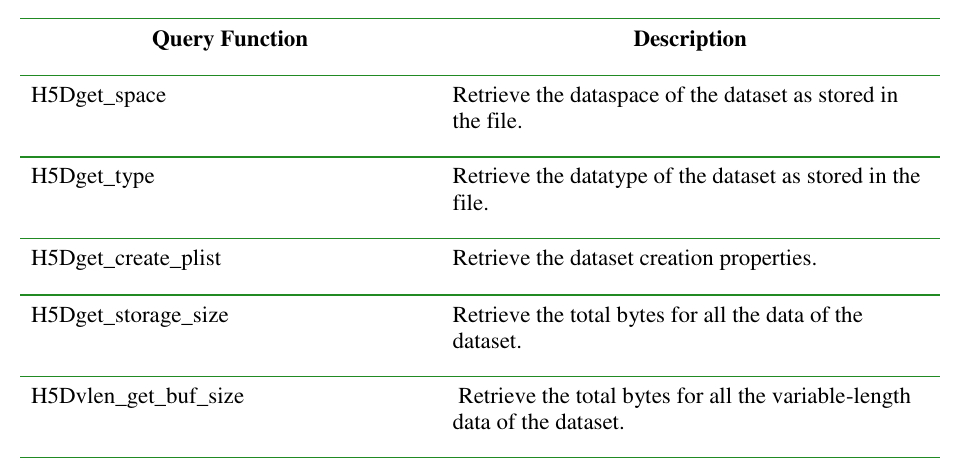
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
|
void CreateFile() {
MatrixD x;
SampleGenerator generator;
int sample_size = 20, n_features = 1000;
VectorD coef, y;
generator.MakeRegression(x, y, coef, sample_size, n_features);
const int MAX = 1e4;
string hdf5_file_address = "extend.h5";
hid_t file_id;
file_id = H5Fcreate(hdf5_file_address.c_str(), H5F_ACC_TRUNC, H5P_DEFAULT, H5P_DEFAULT);
hsize_t x_dims[2] = {sample_size, static_cast<hsize_t>(n_features)}, max_dims[2] = {H5S_UNLIMITED, H5S_UNLIMITED};
hsize_t chunk[2] = {MAX, static_cast<hsize_t>(n_features)};
hid_t X_space, X_id, Y_space, Y_id, X_dcpl, Y_dcpl, X_mem_space, Y_mem_space;
int rank = 2;
X_space = H5Screate_simple(rank, x_dims, max_dims);
X_dcpl = H5Pcreate(H5P_DATASET_CREATE);
H5Pset_chunk(X_dcpl, 2, chunk);
X_id = H5Dcreate2(file_id, "/X", H5T_NATIVE_DOUBLE, X_space, H5P_DEFAULT, X_dcpl, H5P_DEFAULT);
H5Pclose(X_dcpl);
H5Dwrite(X_id, H5T_NATIVE_DOUBLE, H5S_ALL, H5S_ALL, H5P_DEFAULT, x.data());
H5Sclose(X_space);
H5Dclose(X_id);
H5Fclose(file_id);
}
void ExtendFile() {
MatrixD x;
SampleGenerator generator;
int sample_size = 1000, n_features = 1000;
VectorD coef, y;
generator.MakeRegression(x, y, coef, sample_size, n_features);
hid_t file_id, faplist_id, X_id, X_space, X_mem_space;
string hdf5_file_address = "extend.h5";
faplist_id = H5Pcreate (H5P_FILE_ACCESS);
H5Pset_fapl_stdio (faplist_id);
file_id = H5Fopen (hdf5_file_address.c_str(), H5F_ACC_RDWR, faplist_id);
X_id = H5Dopen2(file_id, "/X", H5P_DEFAULT);
X_space = H5Dget_space(X_id);
hsize_t X_dims[2], x_start[2], x_count[2];
x_start[1] = 0;
x_count[1] = n_features;
H5Sget_simple_extent_ndims(X_space);
H5Sget_simple_extent_dims(X_space, X_dims, NULL);
x_start[0] = X_dims[0];
X_dims[0] += sample_size;
H5Dset_extent(X_id, X_dims);
X_space = H5Dget_space(X_id);
x_count[0] = sample_size;
int rank = 2;
X_mem_space = H5Screate_simple(rank, x_count, NULL);
H5Sselect_hyperslab(X_space, H5S_SELECT_SET, x_start, NULL, x_count, NULL);
H5Dwrite(X_id, H5T_NATIVE_DOUBLE, X_mem_space, X_space, H5P_DEFAULT, x.data());
H5Sclose(X_space);
H5Sclose(X_mem_space);
H5Dclose(X_id);
H5Fclose(file_id);
}
void ExtendFile2() {
MatrixD x;
SampleGenerator generator;
int sample_size = 1000, n_features = 1000;
VectorD coef, y;
hid_t file_id, faplist_id, X_id, X_space, X_mem_space;
string hdf5_file_address = "extend.h5";
faplist_id = H5Pcreate (H5P_FILE_ACCESS);
H5Pset_fapl_stdio (faplist_id);
file_id = H5Fopen (hdf5_file_address.c_str(), H5F_ACC_RDWR, faplist_id);
int time = 13000;
for (int i = 0; i < time; i++) {
generator.MakeRegression(x, y, coef, sample_size, n_features);
X_id = H5Dopen2(file_id, "/X", H5P_DEFAULT);
X_space = H5Dget_space(X_id);
hsize_t X_dims[2], x_start[2], x_count[2];
x_start[1] = 0;
x_count[1] = n_features;
H5Sget_simple_extent_ndims(X_space);
H5Sget_simple_extent_dims(X_space, X_dims, NULL);
x_start[0] = X_dims[0];
X_dims[0] += sample_size;
H5Dset_extent(X_id, X_dims);
X_space = H5Dget_space(X_id);
x_count[0] = sample_size;
int rank = 2;
X_mem_space = H5Screate_simple(rank, x_count, NULL);
H5Sselect_hyperslab(X_space, H5S_SELECT_SET, x_start, NULL, x_count, NULL);
H5Dwrite(X_id, H5T_NATIVE_DOUBLE, X_mem_space, X_space, H5P_DEFAULT, x.data());
}
H5Sclose(X_space);
H5Sclose(X_mem_space);
H5Dclose(X_id);
H5Fclose(file_id);
}
void ExtendFile_mpi() {
MatrixD x;
SampleGenerator generator;
int sample_size = 500, n_features = 1000;
VectorD coef, y;
generator.MakeRegression(x, y, coef, sample_size, n_features);
int mpi_size, mpi_rank;
MPI_Comm comm = MPI_COMM_WORLD;
MPI_Comm_rank(comm, &mpi_rank);
MPI_Comm_size(comm, &mpi_size);
string file_address = "extend.h5";
hid_t file_access, file_id, plist_id;
file_access = H5Pcreate(H5P_FILE_ACCESS);
H5Pset_fapl_mpio(file_access, comm, MPI_INFO_NULL);
file_id = H5Fopen(file_address.c_str(), H5F_ACC_RDWR, file_access);
H5Pclose(file_access);
hid_t X_id, X_space, mem_space;
X_id = H5Dopen2(file_id, "/X", H5P_DEFAULT);
X_space = H5Dget_space(X_id);
hsize_t X_dims[2], x_start[2], x_count[2], offset[2];
x_start[1] = 0;
x_count[1] = n_features;
H5Sget_simple_extent_ndims(X_space);
H5Sget_simple_extent_dims(X_space, X_dims, NULL);
x_start[0] = X_dims[0] + mpi_rank * sample_size;
X_dims[0] += sample_size * mpi_size;
H5Dset_extent(X_id, X_dims);
X_space = H5Dget_space(X_id);
x_count[0] = sample_size;
int rank = 2;
mem_space = H5Screate_simple(rank, x_count, NULL);
H5Sselect_hyperslab(X_space, H5S_SELECT_SET, x_start, NULL, x_count, NULL);
// MPI_Barrier(comm);
plist_id = H5Pcreate(H5P_DATASET_XFER);
H5Pset_dxpl_mpio(plist_id, H5FD_MPIO_COLLECTIVE);
H5Dwrite(X_id, H5T_NATIVE_DOUBLE, mem_space, X_space, plist_id, x.data());
// MPI_Barrier(comm);
H5Dclose(X_id);
H5Sclose(mem_space);
H5Sclose(X_space);
H5Pclose(plist_id);
H5Fclose(file_id);
}
|